Breakthrough in Developmental Biology: 3D Atlas of Fruit Fly Cells Offers New Insights
BEIJING -- In a groundbreaking achievement, researchers have developed a comprehensive three-dimensional spatiotemporal multi-omics atlas of single cells throughout the entire developmental cycle of fruit flies. This innovative atlas provides invaluable molecular-level insights into the biological processes driving development, making it a significant advancement in understanding developmental defects and related disease mechanisms.
The study represents a collaborative effort between BGI Research, located in Hangzhou, and the Southern University of Science and Technology in Shenzhen. It has been published in the esteemed journal Cell, highlighting the importance of its findings in the field of developmental biology.
Describing animal development as an intricately coordinated process, scientists emphasize how genes and cells work together with exceptional spatiotemporal precision. Using fruit flies, which are widely recognized as a model organism in biological research, the researchers explored the fruit fly’s developmental cycle, which consists of four distinct phases: egg, larva, pupa, and adult. This progression can be metaphorically compared to a meticulously staged “living theater production,” where each cell's timing for entry, its spatial arrangement, and its transformation into specialized cell types are finely regulated by genetic “scripts.”
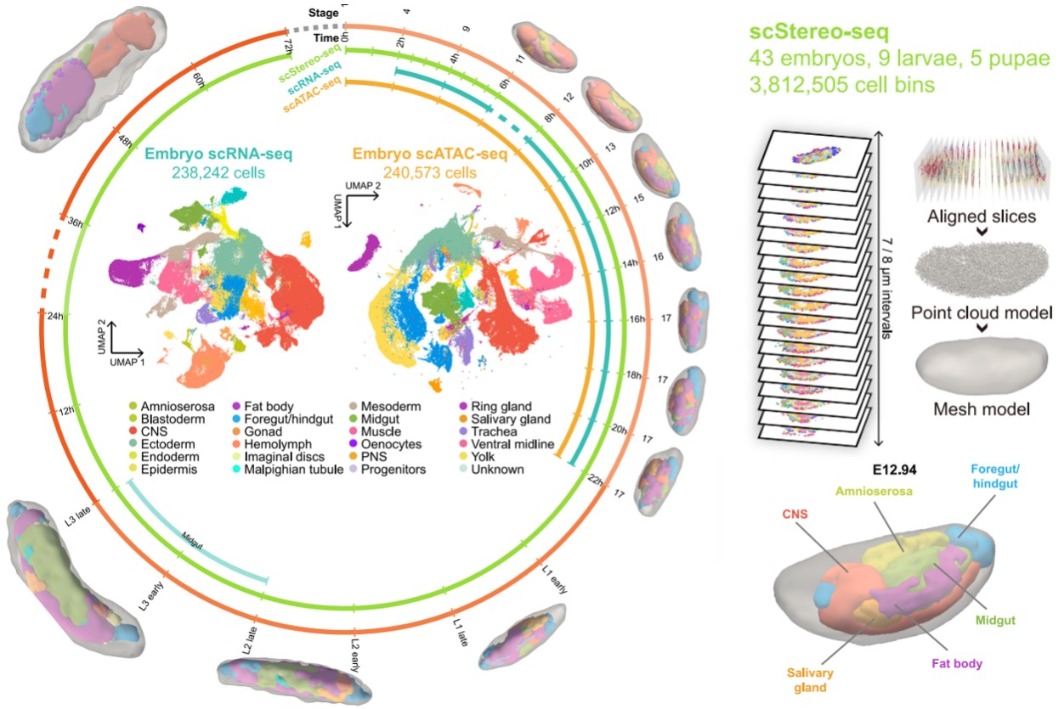
The research team leveraged BGI’s proprietary technologies to perform intensive sampling of fruit fly embryos at intervals ranging from 30 minutes to two hours. This was complemented by systematic sampling during the larval and pupal stages at critical developmental time points. Such a rigorous strategy yielded an enormous dataset comprising over 3.8 million spatially resolved single-cell transcriptomes, encompassing the entire life cycle of the fruit fly.
Employing Spateo, a sophisticated algorithmic tool designed for spatiotemporal analysis, the researchers were able to reconstruct a high-resolution 3D model that accurately illustrates the spatial dynamics of tissue morphology and gene expression. By integrating this diverse range of data, they created a differentiation trajectory map that sheds light on the fundamental molecular mechanisms that govern cell fate decisions.
According to Wang Mingyue, a co-first author of the study, “Cells from different germ layers follow distinct differentiation paths. Transcription factors act as ‘cellular directors,’ orchestrating differentiation by activating or repressing genes to assign specific roles to cells.” Wang’s insights reveal that numerous previously unknown transcription factors were identified, which may play crucial roles in the development of the nervous system, gut, and endocrine systems.
This research is particularly promising given that approximately 70 percent of human disease-related genes have counterparts in fruit flies. Wang emphasizes that this research offers a powerful reference point for investigating human developmental diseases and opens new avenues for biomedical research, potentially leading to innovative treatments and therapies for various conditions.